Doi: 10.3389/fmicb.2022.932840
As the temperature descends, the enchantment of a warm hot spring becomes irresistible, beckoning us to indulge in the comforting warmth of immersion. However, you might not have been aware that amidst the cozy embrace of these hot springs, a group of tiny and mysterious microorganisms silently becomes our bathing companions. These microorganisms are known as thermophilic cyanobacteria. Flourishing in the extreme temperatures (45-73°C) of Asian non-acidic hot springs, they actively engage in photosynthesis and multiply, showcasing remarkable adaptability. Undoubtedly, they stand out as the most representative microorganisms in hot springs.
Thermophilic cyanobacteria possess highly distinctive physiological functions, thriving in elevated temperatures to sustain normal cellular growth and metabolic activities, while experiencing inhibition at regular temperatures. Their enzymes showcase remarkable heat stability, making them an excellent source of thermostable enzymes. Additionally, thermophilic cyanobacteria play a pivotal role as the primary producers in hot spring ecosystems, converting sunlight into energy through photosynthesis and releasing oxygen in the process. This photosynthetic capability serves as a vital source of energy and oxygen for other microorganisms in the entire hot spring ecosystem. In recent years, with the rapid advancement of biotechnology, these hot spring cyanobacteria have emerged as crucial tools for bioenergy production and environmental restoration. Therefore, a thorough analysis of their genomes not only enhances our understanding of their unique physiological functions and evolutionary history but also contributes to the conservation and sustainability of ecosystems, as well as the application and development of biotechnology.
Among thermophilic cyanobacteria, the genus Thermosynechococcus dominates the microbial mats in Asian non-acidic hot springs. To unravel the scientific mysteries inherent in these microorganisms, Drs. Hsiu-An Chu and Chih-Horng Kuo, together with their research team at the Institute of Plant and Microbial Biology, Academia Sinica, conducted a comprehensive genome analysis of Thermosynechococcus sp. CL-1. This strain, collected by Dr. Hsin-Ta Hsueh (National Cheng-Kung University) from the Chin-Lun hot spring in Taitung (pH 9.3, 62°C) in 2020, exhibited highly different chromosomal organization, gene content, and divergent phylogenetic relationships compared to other species from the same genus. Later in 2022, the genomic study of another strain, Thermosynechococcus sp. TA-1, collected by Dr. Jyh-Yih Leu (Fu-Jen Catholic University) from the Tai-An hot spring in Miaoli (pH 7.5, 40-63°C), was conducted and compared with CL-1. Additionally, a compilation of Thermosynechococcus strains with complete genome sequences from GenBank was utilized for further comparisons.
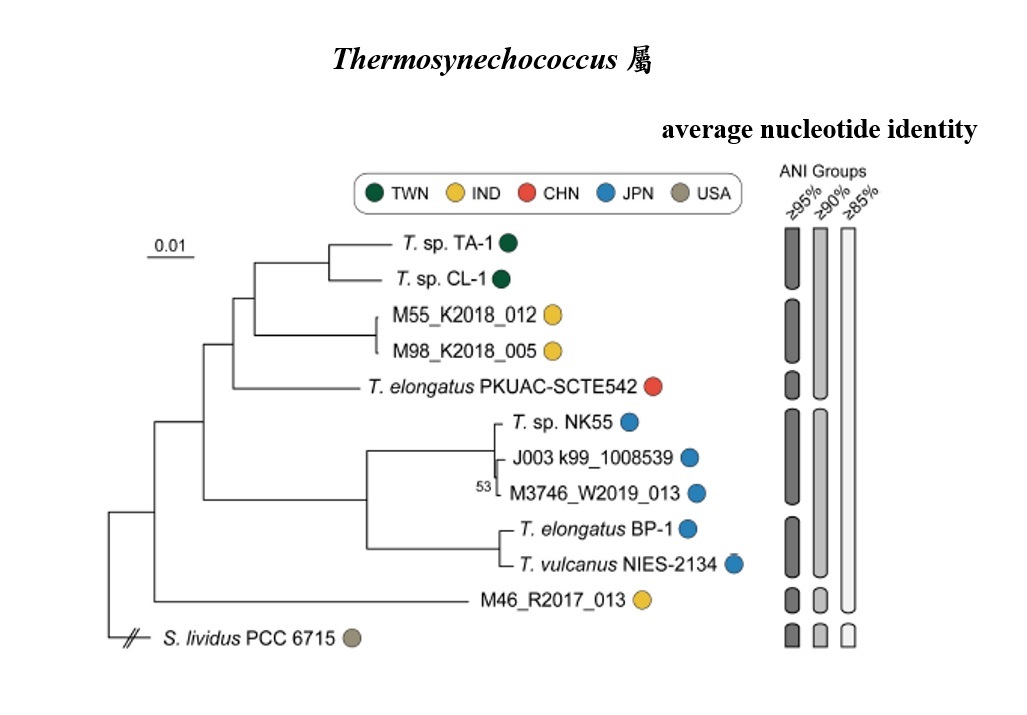
Through genomic analysis, the research team discovered that the average nucleotide identity (ANI) between TA-1 and CL-1 is approximately 97.0%, which is above the 95% threshold suggested for delineating bacterial species.
This signifies that the Thermosynechococcus strains TA-1 and CL-1 from Taiwan belong to a novel species-level taxon. In contrast, the ANI between these Taiwanese thermophilic cyanobacteria and those from other countries fall below the 95% threshold, indicating that TA-1 and CL-1 represent a new species exclusive to Taiwan.
Doi: 10.3389/fmicb.2022.932840
Another intriguing observation is that the evolution and genomic diversity of these cyanobacteria seem closely tied to their geographical locations. For instance, strains from Taiwan (TA-1 and CL-1), India (M55 and M98), and China (E542) form one clade, while all the Japanese strains constitute another clade. They show a distinct separation from the hot spring cyanobacterium collected from the United States (S. lividus PCC 6715), indicating a more distant evolutionary relationship.
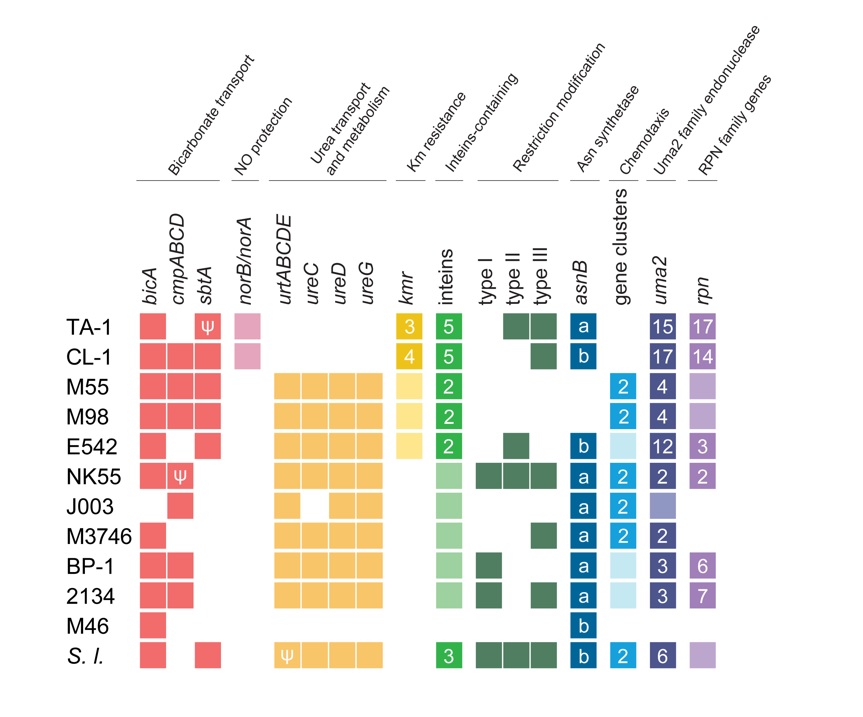
Furthermore, comparative analysis reveals distinct features in the genomes of Taiwanese hot spring cyanobacteria strains TA-1 and CL-1.
Notably, TA-1 and CL-1 share genes related to nitric oxide protection and kanamycin resistance. Conversely, these strains lack genomic components associated with urea transport and chemotaxis. This underscores the evolutionary adaptation of Taiwanese hot spring cyanobacteria strains, suggesting either gene gains by horizontal transfer or gene losses through reductive evolution, which helped these strains to thrive in Taiwan’s unique hot spring ecosystem.
Doi: 10.3389/fmicb.2022.932840
The study of thermophilic cyanobacteria opens a fascinating window into the world of microorganisms, providing profound insights into life’s ability to thrive in extreme environments. The discovery of unique genes not only deepens our understanding of life but also becomes a pivotal resource for advancing biotechnology and ecological research. In conclusion, hot springs are repositories teeming with extraordinary life wonders. Anticipation builds for future investigations that will unravel more mysteries concealed within these minute organisms, offering valuable revelations for ecology, biotechnology, and beyond. Each hot spring in Taiwan conceals a multitude of tales involving these microorganisms, eagerly awaiting revelation in the future.
【Suggested Reference】
Yen-I Cheng, Yu-Chen Lin, Jyh-Yih Leu, Chih-Horng Kuo*, Hsiu-An Chu*
Comparative analysis reveals distinctive genomic features of Taiwan hot-spring cyanobacterium Thermosynechococcus sp. TA-1.
Front Microbiol (2022) 13:932840.
🥒https://doi.org/10.3389/fmicb.2022.932840
Yen-I Cheng, Lin Chou, Yi-Fang Chiu, Hsin-Ta Hsueh, Chih-Horng Kuo*, Hsiu-An Chu*
Comparative genomic analysis of a novel strain of Taiwan hot-spring cyanobacterium Thermosynechococcus sp. CL-1
Front Microbiol (2020) 11:82.
🥒https://doi.org/10.3389/fmicb.2020.00082
