The 2022 outbreak of monkeypox (mpox) was declared as a WHO public health emergency of international concern, following that of the coronavirus pandemic. There were over 80,000 confirmed cases of monkeypox worldwide from May to the end of December 2022. As of July 3, 2023, there have been a total of 221 confirmed cases of monkeypox in Taiwan. Typical symptoms observed include fever and skin manifestations such as rashes, spots, blisters, and pustules. Did you know that this alarming monkeypox virus actually belongs to the “giant viruses”?
in marine ecosystems
The term “giant viruses” already gives a hint that these viruses are vastly different from the viruses we are familiar with. Not only are they gigantic in size, but they also possess large complex genomes.
Giant viruses comprise the phylum Nucleocytoviricota recently established by the International Committee on Taxonomy of Viruses (ICTV). Traditionally, viruses have been regarded as tiny and elusive microbes that parasitize host cells. However, some remarkable discoveries came from studies on this group of giants. Unlike the majority of viruses, which are typically smaller than 100 nanometers (nm), giant viruses can exceed 200 nm and even reach sizes over 1,000 nm, comparable to some bacteria. What’s even more astonishing is that giant viruses possess double-stranded DNA genomes containing hundreds to over two thousand genes, surpassing the gene repertoire of some bacteria. Giant viruses have been found to infect a wide range of eukaryotic organisms, including various animals, algae, amoebae, and flagellates (Table 1).
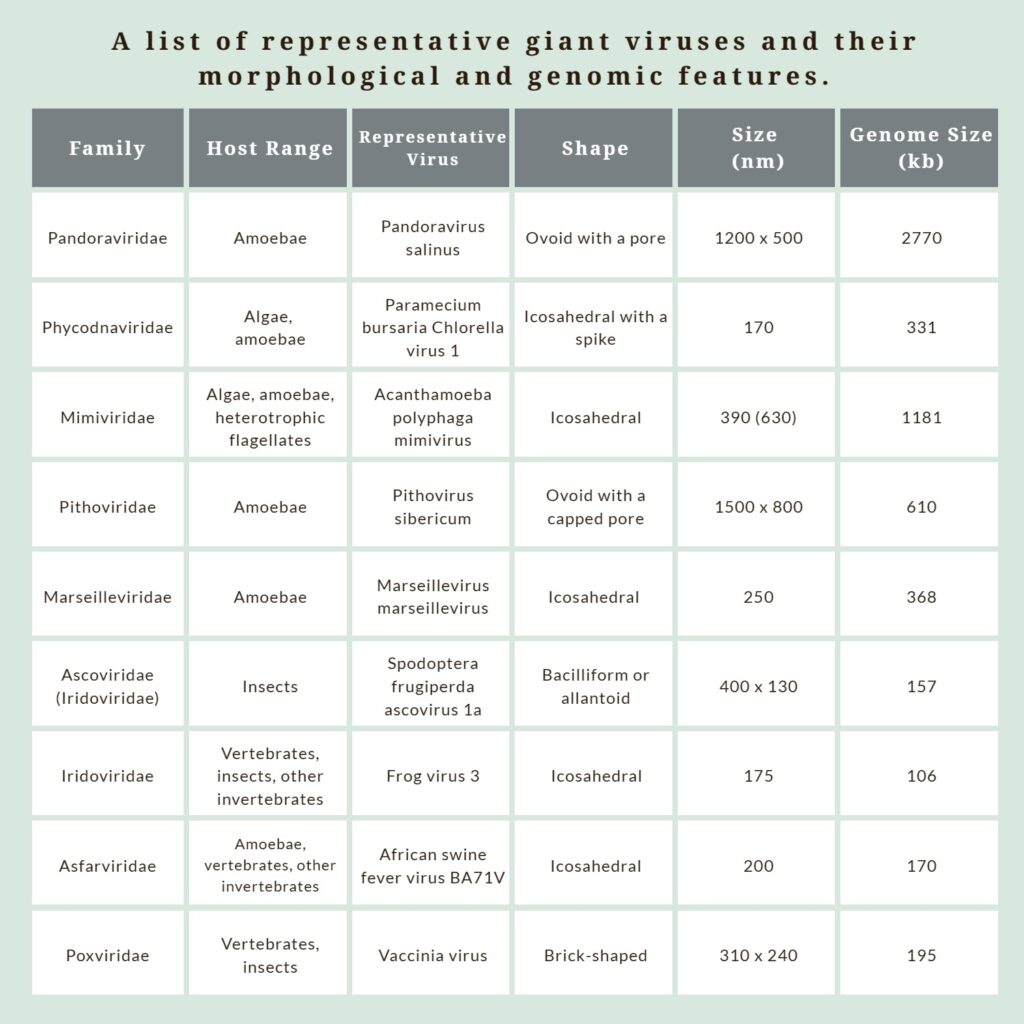
and representative viruses
The reason why the genomes of giant viruses became so large remains a major mystery in the field of evolutionary biology.
In the past, our understanding of giant virus evolution relied primarily on a few widely shared core genes to infer the evolutionary relationships between different viruses and define their taxonomic groups such as classes, orders, and families. However, through whole-genome analysis, we can quantify the degree of shared homologous genes among giant viruses. A recent study conducted by Chuan Ku’s Lab at the Institute of Plant and Microbial Biology, Academia Sinica, has shown that giant virus lineages that are closer in the core gene tree may not necessarily share a higher number of genes. This phenomenon is particularly evident at higher taxonomic levels. For example, the two families in the class Pokkesviricetes, Asfarviridae and Poxviridae, exhibit lower gene sharing between them compared to each of them with some other families. The study further demonstrates that the host diversity of different families correlates with their intrafamilial gene repertoire variation. Viruses from different families but infecting a similar type of hosts (algae, vertebrates, or insects) tend to share more genes, such as those encoding ion channels or animal immune response proteins. By exerting selection or providing a pool of genes that can be transferred, the hosts, their environment, and their associated microbes shape the gene contents of giant viruses and potentially play a crucial role in viral genome gigantism and the origin of giant viruses.
Reference:
Tsu-Wang Sun*, Chuan Ku (2021) Unraveling gene content variation across eukaryotic giant viruses based on network analyses and host associations. Virus Evolution. https://doi.org/10.1093/ve/veab081
When a virus infects a host, the fusion between virus and host membranes plays a crucial role in releasing viral genetic material and initiating infection. While most viruses rely on a single protein for membrane fusion, giant viruses like poxviruses employ an entry/fusion complex (EFC) composed of multiple proteins to mediate this process. To unravel the origin of the poxviral EFC, the labs of Chuan Ku (Institute of Plant and Microbial Biology) and Wen Chang (Institute of Molecular Biology) conducted a comprehensive genomic survey of giant viruses. Their findings revealed that the two major EFC protein families are widely distributed across giant viruses. They discovered that each of these two protein families is structurally conserved, traces its origin to the root of Nucleocytoviricota, then passed down to the major descendant lineages of Nucleocytoviricota, and is retained in most giant viruses that are known to infect animals and amoebozoans. These findings indicate that the EFC proteins represent a potentially conserved mechanism for cell entry in diverse giant viruses. The work expands our knowledge gained from model poxvirus systems and provides fresh insights into the origin, evolution, and biology of these enigmatic giant viruses.
Reference:
Sheng Kao, Chi-Fei Kao, Wen Chang, Chuan Ku* (2023) Widespread distribution and evolution of poxviral entry-fusion complex proteins in giant viruses. Microbiology Spectrum. https://journals.asm.org/doi/10.1128/spectrum.04944-22
The discoveries of giant viruses, particularly those with greater impacts on the human society, have raised massive attention and concern. Examples include monkeypox virus and African swine fever virus, which not only have a high infectivity but also pose significant threats to human health and food safety. Furthermore, the existence of giant viruses challenges traditional view of viruses and opens up a new area of research. Scientists are actively exploring these enormous and complex viruses, aiming to unravel their origins, evolution, and interactions with hosts. Further research on viral genome gigantism and the role of hosts in this process will contribute to a deeper understanding of the mysteries of the viral world and provide new perspectives on the evolution of life.
